A latent mixture model for heterogeneous causal mechanisms in Mendelian randomization
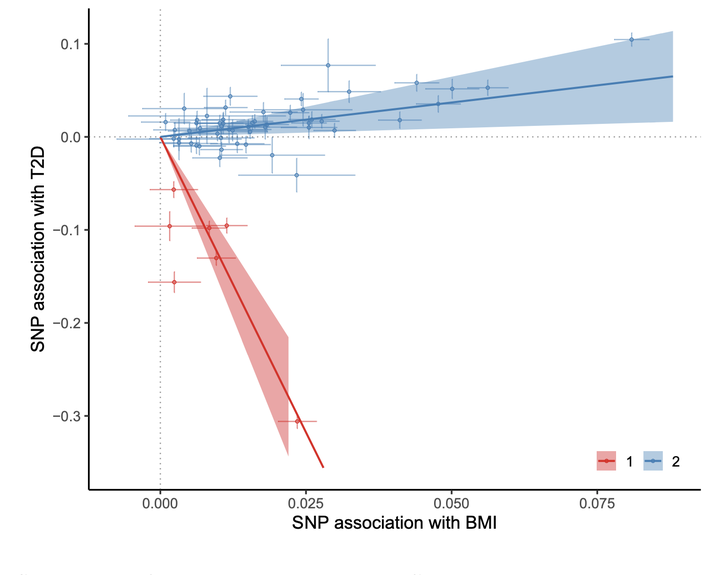 Scatterplot of BMI-T2D data with MC-EM parameter estimates.
Scatterplot of BMI-T2D data with MC-EM parameter estimates.
Abstract
Mendelian Randomization (MR) is a popular method in epidemiology and genetics that uses genetic variation as instrumental variables for causal inference. Existing MR methods usually assume most genetic variants are valid instrumental variables that identify a common causal effect. There is a general lack of awareness that this effect homogeneity assumption can be violated when there are multiple causal pathways involved, even if all the instrumental variables are valid. In this article, we introduce a latent mixture model MR-PATH that groups instruments that yield similar causal effect estimates together. We develop a Monte-Carlo EM algorithm to fit this mixture model, derive approximate confidence intervals for uncertainty quantification, and adopt a modified Bayesian Information Criterion (BIC) for model selection. We verify the efficacy of the Monte-Carlo EM algorithm, confidence intervals, and model selection criterion using numerical simulations. We identify potential mechanistic heterogeneity when applying our method to estimate the effect of high-density lipoprotein cholesterol on coronary heart disease and the effect of adiposity on type II diabetes.